Top Links
Journal of Plant Sciences and Crop Protection
ISSN: 2639-3336
Transgenic Expression of Sugarcane Mosaic Virus VPg in Maize Inbred Line CML444 Confers Resistance to Maize Lethal Necrosis Disease
Copyright: © 2023 Obara Justus Anyieni. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
Maize is the most important crop in Kenya and parts of Sub-Saharan Africa. However, yields are below potential due to abiotic and biotic constraints. One of the major biotic concerns is maize lethal necrosis disease (MLN), which causes up to 100% yield losses. MLN is caused by the synergistic infection of two viruses, Maize chlorotic mottle virus (MCMV) and a potyvirus, commonly Sugarcane mosaic virus (SCMV). Because MLN is dependent on synergism, reduction of infection by either virus is expected to confer resistance or tolerance to MLN. Proteins P1, HC-Pro and VPg are essential for SCMV replication and movement. Pathogen-derived resistance has been used to design antiviral resistance in plants. Here, we hypothesized that transgenic expression of SCMV P1, HC-Pro or VPg confers resistance to SCMV and thus, to MLN. To test this hypothesis, we transformed maize inbred line CML444 with SCMV P1, HC-Pro or VPg genes; confirmed the presence of the transgene in T1 plants and evaluated T2 for MLN resistance using a detached leaf assay. Whole plant assays were not possible due to the legal restrictions of genetically modified plants in Kenya. MLN severity was evaluated on a scale of 1-5 using the chart developed by CIMMYT. Leaves from VPg transgenic plants recorded a severity score of 1.44 representing no MLN. In contrast, leaves from P1 and the HC-Pro transgenic plants had excessive chlorosis with a score of 4.0 and 4.1, respectively, while the susceptible control was completely chlorotic with a severity score of 5.0. Based on the area under disease progress curve, the VPg, HC-Pro, and P1 transgenic had 6.44%, 22.43%, and 17.48%, respectively, while the susceptible control had 23.13%. Analysis of variance revealed that the area under disease progress curve and MLN severity scores were significantly different across the transgenes, with transgenic expression of VPg providing the most protection against MLN. These results show that MLN management can be improved through gene silencing induced by transgenic expression of SCMV VPg.
Keywords: VPg; Transgenic Maize; Host Induced Gene Silencing; Small Interfering RNAs; Transgenic Resistance.
Emergence and re-emergence of pests and diseases has caused significant reduction in maize production in the last decade. Maize lethal necrosis disease (MLN) re-emergence ravaged crops threatening livelihoods in the East and Central Africa [1]. Two viruses, Maize chlorotic mottle virus (MCMV) in combination with several species in the family Potyviridae, including Sugarcane mosaic virus (SCMV), Maize dwarf mosaic virus (MDMV) or Wheat streak virus (WSMV) cause MLN [2]. However, in East and Central Africa, MLN is caused by co-infection of MCMV and (SCMV). In co-infected plants, MCMV accumulates to higher levels, compared to single MCMV infection [3]. Some of the notable symptoms during a severe MLN are leaf yellowing and plant drying, premature plant death, stunted growth and low grain setting in the infected plants [4].
A number of measures are in place to manage MLN including cultural, chemical and physical treatment of the infected plant materials [1]. However, these methods are not economic and environmental friendly [5]. Therefore, a robust, environmental friendly and sustainable tool would be the future solution [6]. New breeding techniques such as gene silencing could offer solutions to virus infections [7]. Gene silencing is one form of pathogen-derived resistance (PDR) where a segment of, or a whole virus gene is integrated into the genome of the host [8]. The integrated nucleic acid activates gene silencing, that results in the formation of small interfering RNAs (siRNAs) complementary to the virus and that reduce establishment of infection by viruses. SiRNAs associate with AGO proteins and program RISC for degradation or translational repression of viral RNA [9,10,11]. Thus, siRNAs derived from the transgenes or transcript areas forming double stranded RNAs (dsRNA) direct silencing of complementary sequences on intruding nucleic acid, such as virus [5].
Previous studies have demonstrated success with transgene activated gene silencing in the management of plant virus diseases [12]. Through transgene activated gene silencing plant viruses such as Pepper mild mottle virus and Plum pox virus were successfully managed [13]. In addition, viral coat protein transgenes were used to suppress SCMV infection in sugarcane or Maize dwarf mozaic virus in maize [14, 15]. Gene silencing initiated by transgenes is specific and shortens breeding time as opposed to the conventional breeding approach [16] Despite this success rate, deployment of gene silencing remains low [5] due to stringent regulations stemming from ethical issues, and inadequate laboratory and technical infrastructures [17, 18].
Observations described above predict that transgene activation of gene silencing against MCMV or SCMV could confer resistance to MLN. Pathogen-derived resistance has been used to design antiviral resistance in plants by activating gene silencing before a virus gets in contact with a plant. Accordingly, this study was based on the hypothesis that transgenic expression of SCMV P1, HC-Pro or VPg genes confers resistance to SCMV and thus to MLN. To test this hypothesis, maize CML444 inbred line was transformed with SCMV P1, HC-Pro or VPg genes and leaves of transgenic plants evaluated. MLN resistance was tested using a detached leaf assay. Results showed that the VPg gene effectively conferred protection against MLN recording a severity score of 1.4 in a 1 to 5 scale. Leaves from transgenic plants expressing P1 and HC-Pro recorded a necrosis score of 4.0 and 4.1, respectively, while the susceptible control was completely necrotic at a score of 5.0. Based on area under disease progress curve, the VPg had the least disease progress (6.4%) followed by the P1, HC-Pro and the susceptible control at 17.5% and 18.3%, 22.4%, respectively. Differences in area under disease progress curve and severity scores were statistically significant. Results described here show that VPg transgene-induced gene silencing could contribute significantly to MLN management.
Twenty-eight day old immature embryo were transformed with plasmids (pMDC32) carrying SCMV P1, VPg or HC-Pro. Genes were separately delivered via Agrobacterium GV3101 [19]. Transformed immature embryos were selected under Kanamycin and transgenic plantlets regenerated in-vitro [20]. In the T1 generation, transgenes were detected and confirmed through PCR and restriction enzyme digestion [15, 21]. Seeds from the T1 transgenics were multiplied in the greenhouse to provide T2 materials for MLN resistance screening.
Virus inoculum was prepared as described [22] with modifications. Plants of susceptible cultivar CML444 co-infected with MCMV and SCMV were used as source. Infected leaves were rinsed 3 times, for 3 min each, in cold autoclaved water. Using mortar and pestle, 100g of tissue were ground in 300 ml of potassium phosphate buffer. The extract was sieved in four-layer sieve sterile cheesecloth and used fresh. These procedures were carried out at 4˚C.
T2 transgenic CML444 seeds were sown in 25×50 cm plastic pots containing peat moss. The pots were kept under anti-insect net, Optinet 50 mesh with (0.26×0.83) pore size in the glasshouse. Maize plants were constantly checked for SCMV and MCMV infection with SCMV ImmunoStrip® KIT (Agdia, USA). At 21 days after sowing a PCR assay was done to confirm presence of the transgenes. Only plants that tested positive for the transgene and were free of SCMV and MCMV were used for MLN resistance screening using a detached leaf assay.
Leaves were collected from 21-day old transgenic plants and washed in cold running tap water to remove adsorbed sand and particulate matter. Leaves were excised with a sterile scalpel into 2 by 2 cm square sections followed by surface sterilized with 50% (v:v) NaClO and 2 drops of Tween20 for 5 minutes under the laminar flow chamber. Leaves were rinsed 3 times, for 3 min each, with cold autoclaved distilled water. Leaf sections were transferred into autoclaved sterile Whatman® filter papers for drying. Microinjuries were inflicted on the leaf sections using a sterile needle.
The MS media was amended with 20 mg/L gibberellic acid, 10 mg/L kinetin and 30% sucrose at 5.8 pH followed by addition of 3 g/L Gelrite. Media was autoclaved at 121˚C for 15 min as described by Tonui [23]. Accurately 30 mL of warm media was dispensed into 100 × 15 mm petri plates under sterile conditions in the laminar flow chamber. Four sterile leaf sections were placed in each plate of MS media with the abaxial surface in contact with media. 20 µL of filter-sterilized inoculum was applied on the injured leaf sections using a sterile ear bud for all the P1, HC-Pro and VPg transgenics and non-transformed leaf sections that were used as susceptible control in the experiments. Plates were sealed in a parafilm and transferred into the growth chamber. Culture conditions were maintained at 26-28°C with 16 hr of light and 8 hr of darkness for 50 days. Fluorescent tubes (PHILIPS-TL-D 18W/54-765) were used for illumination. Occasionally, petri plates with excess water were opened in the laminar flow chamber to allow for escape of excess water. The experiment was laid in a completely randomized design in three replicates.
The detached leaf sections were monitored daily for manifestation of chlorosis. Starting at 5 days after inoculation, scoring of chlorosis was done every 5 days for 50 days. A numerical score was used according to the MLN-severity chart with a scale of 1 to 5 [24]. In this scale, 1 represents no maize lethal necrosis disease symptoms, 2 means fine chlorotic streaks, 3 represents chlorotic mottling throughout a leaf, 4 means excessive chlorotic mottling and dead heart; and 5 is for complete plant necrosis). Area under disease progress curve (AUDPC) was computed as described [24, 25].
Data on MLN severity in each plate was converted to the area under the disease progress curve following the formula described by CIMMYT [25].
Where n=total number of disease observations, t=time (day) of each disease assessment, yi=disease severity expressed as a proportion at the ith assessment, yi+1=disease severity expressed as proportion at subsequent assessment date (day) (i + 1), ti= time during the ith assessment and ti+1= the second consecutive disease assessment date (day).
The severity and AUDPC data were subjected to analysis of variance (ANOVA) using SAS version 9.4 (SAS Institute, 2001). The analysis followed an appropriate model for complete randomized design (CRD) using proc glm procedure of SAS.
where, Yijk = observation of the experimental units, µ = overall mean, Gi= effects due to ith genes, and Ꜫijk = residual.
Upon significance of the main effects (genes), mean separation was conducted using Least significance difference (LSD) procedure at a p ≤ 0.05 level of significance for each disease severity and AUDPC estimates.
Throughout the assay period, disease severity progressed rapidly in all genotypes tested except on leaves from the VPg transgenic plants (Figure 1). From 5 to 20 days post inoculation, the VPg, transgenic had a severity score of 1.0, 1.0, 1.0, and 1.0. In contrast, on leaves of transgenic plants expressing P1 severity increased from 1.8, 2.0, 3.0 to 3.67; from 1.8, 2.5, 31 and 4.0 on leaves of plants expressing HC-Pro and from 2.25, 3.5, 4.5 and 5.0 on the susceptible control. For the rest of the assay period (between 30 and 50 days post inoculation) the VPg transgenic recorded severity values of 1.4, 1.5, 1.6 and 1.6. The P1 remained at values between 4.0, and 4.08, and HC-Pro remained between 4.0, and 4.17, while the susceptible control reached values of 5.0 (Figure 2). At the end of the 50-day evaluation period, the VPg transgenic had the minimum disease score at 1.6 indicating no MLN. The P1 at and HC-Pro had a final score of 4.0 and 4.1, respectively, indicating excessive chlorosis while the susceptible control was completely necrotic with a score of 5.0. (Figures 1 and 2). Similarly, VPg transgenics recorded the lowest value of area under disease progress curve, followed by the P1 and HC-Pro transgenics while the susceptible control had the highest value among the tested materials at the end of the 50-day assay period (Figure 3).
The VPg transgenics had the least area under disease progress as compared to the HC-Pro, P1 transgenics and the susceptible control, which is predicted to derive from successful activation of gene silencing gainst SCMV VPg. However, the VPg transgenic proteins also might inhibit the formation of VPg-maize Elongin C (ZmElc) proteins complexes [26], thus contributing to a reduction in SCMV replication. During a successful SCMV invasion, the VPg-ZmElc protein complex functions as replication primers an event that increases MLN severity [27].
The three proteins, P1, HC-Pro and VPg are important in the formation of virus replication compartments (VRC), a process that converts cell membranes into miniorganells for viruses to replicate that house viral RNA and host proteins necessary for replication [28]. It is also likely that silencing VPg gene was more effective in antagonizing the formation of virus replication compartments (VRCs) as compared to the HC-Pro and P1 transgenics [29]. Alternatively, or in addition, HC-Pro and P1 proteins are less stable than VPg. However, it is also possible that the effects are mediated by small RNAs derived from the transgenes instead of at the protein level.
During MLN pathogenesis, the SCMV HC-Pro and VPg proteins bind the maize ferredoxin-5 (FdV) causing a disruption of its import into the maize bundle-sheath cell (BSC) of the chloroplast where it is involved in electron [30]. An influx of electrons in leaves reduces NADP+ via a ferrodoxin NADP oxidoreductase (FNR) to create energy. Inadequate or completely no arrest of these proteins in the HC-Pro, P1 and the control experiments might have altered the chloroplast transport via the formation of the P1/HC-Pro-FdV complexes, which led to low ATP synthesis less what is required during the Calvin Cycle [3]. This could be the underlying reason for the observed leaf chlorosis in the rest of the leaf sections as opposed to the VPg transgenic. Leaf chlorosis is normally the main cause for reduced rates of photosynthesis in MLN infections due to sub-optimal chlorophyll synthesis [30, 31].
Results described here are in agreement with several published results on the use pathogen-derived resistance trough activation of gene silencing. Maize Dwarf Mosaic Virus (MDV) causes leaf chlorosis and death in maize and sorghum leading to significant losses. Resistance was generated by introducing a 150-base pair segment of protein P1 in a hairpin to form dsRNA. The transgene activates gene silencing and three independent transgenic lines were found to be immune [32]. Transgenic expression of SCMV CP in sugarcane resulted in plants immune to SCMV [14]. Resistance to Wheat streak mosaic virus (WSMV) was generated by expressing the entire NIb and CP as transgenes [5].
Post-genetic modification screening for disease resistance is important in obtaining better-adapted crops. A reliable disease resistance screening approach for separation of susceptible and resistant lines is recommended [33]. Routinely, screening for disease resistance in crops takes place in the greenhouse or fields. However, these two screening approaches possess enormous challenges. Triggering disease pressure through the natural environment is unpredictable. Occurrence of simultaneous non-target infections is possible in the field/greenhouse, which is likely to compromise results. Scouting of viral diseases based on eye inspection is not reliable. Greenhouse screening is reliant on seedling age, inoculum quality, quantity, and environmental conditions, which further confines this approach. The process is laborious and human error prone Yavis [33] in addition high cost, inability to test multiple disease interactions and possibilities of spillover [34].
Detached leaf assay (DLA) would therefore, safeguard against accidental disease or transgene ‘spill-over’ [35]. The method confers convenience when testing for large numbers of genotypes, in screening for multiple pathogens and multiple pathogen interactions. The DLA method requires less screening space, thus reducing experimental costs [36]. Moreover, DLA has demonstrated correlations with the greenhouse results with successful screening of pathogens such as fusarium head blight, angular leaf spot disease and MLN [37, 38, 39].
The absence of MLN in leaves transformed with VPg indicate a successful suppression of SCMV infection due to transgenic VPginduced gene silencing. This study demonstrated that management of MLN through transgene-induced gene silencing is possible. Therefore, transgenic-mediated resistance through RNA could be integral in providing timely solutions towards MLN management. We anticipate for full plant assays under greenhouse and field conditions in our future studies.
The authors reported no potential conflict of interest.
Center of Excellence for Sustainable Agriculture & Agribusiness Management (CESAAM), Egerton University.
OJA conceived the study and executed experiment. HGR provided the plasmids carrying VPg, HC-Pro and P1. OJA, HGR and SM contributed to setting up experiments. RM, MO, MC analyzed the data and drafted the manuscript. OJA and HGR wrote the final manuscript. All authors have read and agreed to the published version of the manuscript.
Plasmids and seed of transgenic plants are available upon request.
We thank the University of Nebraska-Lincoln Center for Virology for their generous donation of the DNA vectors. We are also thankful to CIMMYT, Kiboko-Kenya for the maize inbred line donation and the Kenyatta University Genetic Transformation Laboratory for granting us the opportunity to conduct the laboratory experiments.
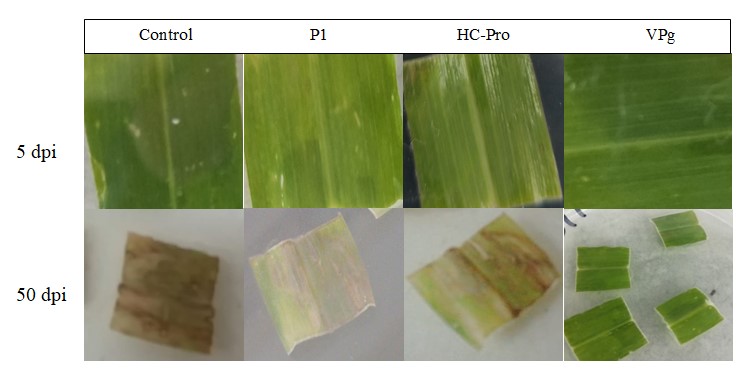 |
| Figure 1: Leaf detached assays showing disease progress in the leaves of maize plants transformed with SCMV VPg, P1 or HC-Pro. Leaves of non-transformed plants were used as susceptible control. Representative pictures were taken at 5 dpi and 50 dpi. The experiment was repeated three times with each petri plate containing 4 leaf sections representing VPg, P1 or HC-Pro transformed plants. |
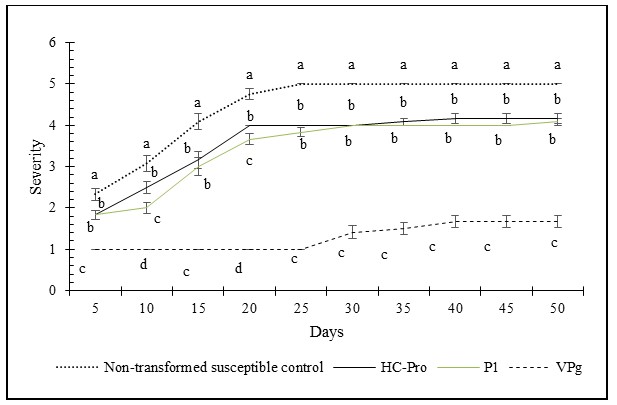 |
| Figure 2: Disease progression on leaf detached assay for a period of 50 days after inoculation. Severity was measured every 5 days in a scale of 1 to 5. C: Non-transformed susceptible control. Each point represents the average and standard error of 4 leaf sections and three biological replicates. At each time point, letters indicate statistical difference (LSD 0.05=2.07) test at p =0.05. |
 |
| Figure 3: Area under disease progress curve at 50 dpi. Values correspond to the last time point in figure 2. Letters indicate statistical difference at p=0.05 (LSD0.05=2.07). |






































