Top Links
Journal of Plant Sciences and Crop Protection
ISSN: 2639-3336
Effect of Hot Water Treatment on DNA Quality of Sweet Orange (Citrus sinensis (L) osbeck) Fruits
Copyright: © 2020 Oladele Oluwole Olakunle. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
In this study, mature sweet orange fruits (amber variety) were separately subjected to hot water treatments: 35 oC for 25 and 30 minutes; 45 oC for 15, 20 and 25 minutes; 48 oC for 10, 15 and 20 minutes; 50 oC for 10, 15 and 20 minutes; 53 oC for 10 minutes and 55 oC for 5 minutes and were thereafter assessed for their DNA quality while unheated fruits served as control. The DNA quality was done by rapid random polymorphic DNA-polymersae chain reaction (RAPD-PCR) on the extracted DNA from each set of treated fruits using the RAPD primer OPR 02 (5’ – CAC AGC TGC C-3’). Agarose gel electrophoresis of PCR products generated after amplification of DNA extracted from hot water –treated fruits showed that high molecular weight (HMW) bands of DNA were detected successfully for control and fruits treated separately at 35 oC for 25 and 30 minutes, 48 oC for 15 and 20 minutes, 45 oC for 20 and 25 minutes and 50 oC for 15 minutes. Thus, this implied that these temperatures – time combinations that gave high molecular bands could be used in extending shelf of orange fruits after harvest.
Keywords: Sweet Orange; Temperature; Exposure Times; Hot Water; DNA
Citrus fruits have several beneficial health and nutritive properties. They are rich in vitamin C and Folic acid, as well as a good source of fiber. They are fat free, sodium and cholesterol free [1]. Besides, they contain vast number of minerals. They may help to reduce the risk of heart diseases and some types of cancer. In short, citrus fruit is enjoyed around the world for its taste, nutritional value and relatively cheap price. Among the citrus species, sweet orange is the most widely grown [2] and according to [3], the recommended varieties of sweet orange fruits in Nigeria include Hamlin, Valencia, Amber sweet, Agege Parson Brown, Umudike, Bende, Etir and Meran. The fruits are either eaten fresh or used for making canned orange juice. Large quantities of sweet orange are used to produce single strength juice, frozen concentrate, rind oil, pectin used in the production of jams and jelly pulp residue which is fed to livestock [4]. The orange juice is also extracted and used for flavorings food items such as salad, desserts, oatmeal and soups.
However, the bulk of sweet orange produced in Nigeria is distributed to prospective consumers through open sale at market stalls and roads stands [5]. In these places, good quality and marketability of the fruits is often lost due to excessive post-harvest spoilage of the fruits which consequently leads to loss of market appeal [6]. Currently, postharvest diseases of perishables are primarily controlled by good sanitation and application of fungicides. Fungicides such as thiabendazole, imazalil and guazatine have been used to control post-harvest fungal diseases of oranges [7]. However, concerns about public health and environmental issues have increased the need for alternatives. In fact, consumers demand and environment concerns in the last few years have led to research on developing or improving non-chemical methods for postharvest protection of horticultural crops. Controlled or modified atmosphere, intermittent warning, illumination, biological control and heat treatment have been recently re-evaluated as options to reduce the use of agricultural chemicals in postharvest treatments.
Heat treatment is usually done in hot water, vapour heat, hot air and recently by radio frequency (RF) heating [8-10]. Investigations on citrus fruit have shown that post-harvest hot water dips (2 – 3minutes at 50 - 53 oC) inhibit or reduce pathogen development [11], improve fruit resistance to chilling injury, effectively reduce water loss in kumquat [12] and greatly increase the efficacy of fungicides applied in postharvest treatments [13]. Unfortunately, many authors who had studied the effects of post-harvest hot water treatments on citrus fruits did so with the sole aim of controlling or reducing post-harvest decay without investigating the effect of such treatments on the DNA quality [11,14,15]. Therefore, the aim of this study was to investigate effect of hot water treatment on DNA quality of sweet orange fruits after harvest.
Fruits of sweet orange (Amber sweet variety) were harvested from the same tree in an orchard, Oluwatuyi, Akure south (7.2146°N and 5.1641°E), taking care to avoid damage by impact. Mature fruits with green rind were selected for uniformity of size and color. They were washed in 1% sodium hypochlorite solution for 1minute, drained and allowed to air dry before hot water treatments.
Fruits were separately immersed in hot water in a water bath fitted with heating elements and mercury in glass thermometer at 35 oC for 25 and 30 minutes; 45 oC for 15, 20 and 25 minutes; 48 oC for 10, 15 and 20 minutes; 50 oC for 10, 15 and 20 minutes; 53 oC for 10 minutes and 55 oC for 5 minutes. Following treatment, the fruits were left to air dry at 28±2 oC for 30 minutes and thereafter assessed for their DNA quality. Untreated fruits served as control./p>
DNA extraction on the skin of the orange fruit was carried out using a ZR plant/seed DNA extraction kit according to manufacturer’s instruction. About 150 mg of the orange skin was added to a ZR BashingBead™ containing 750μl of the lysis solution. The skin was then homogenized using a mini bead beater for minutes. After homogenization, the tube was centrifuged at 10,000xgfor 1 minute. 400 μl of the supernatant was transferred into a Zymo-Spin™ IV Spin Filter in a Collection Tube and centrifuged at 7,000 rpm for 1 minute. 1, 200 μl of Plant/Seed DNA Binding Buffer was then added to the filtrate in the Collection Tube. This mixture was transferred to a Zymo-Spin™ II Column in a Collection Tube, and centrifuged at 10,000 x g for 1 minute. The flow through from the Collection Tube was discarded, and 500 μl of Plant/Seed DNA Wash Buffer was added to the Zymo-Spin™ II Column in a new Collection Tube and centrifuged at 10,000 x g for 1 minute.
After repeating the wash step, the column was transferred to a clean 1.5 ml micro centrifuge tube and 100 μl of sterile water was added directly to the column matrix after which the tubes was centrifuged at 10,000 x g for 30 seconds to elute the DNA. Finally, the eluted DNA was filtered using a Zymo-Spin™ IV-HRC Spin Filter into a 1.5 ml micro centrifuge tube and centrifuged at 10,000 x g for 30 seconds. Rapid random polymorphic DNA-Polymersae chain reaction (RAPD-PCR) was carried out on the extracted DNA sample using the RAPD primer OPR 02 (5’ – CAC AGC TGC C-3’). The PCR reaction was carried out in a 25μl reaction mixture containing 1X PCR buffer (Solis Biodyne), 2.5mM Magnesium Chloride, 0.2mM of each dNTP, 40 pMol of primer, 1U Taq DNA polymerase, 10-200ng of DNA, and sterile deionized water was used to make up the reaction mixture. Amplification was carried out in an Eppendorf Nexus thermal cycler using the following cycling parameters; an initial denaturation at 95 oC for 5 minutes which was followed by 40 consecutive cycles of 95 oC for 1 minute, 30 oC for 1 minute and 72 oC for 2 minutes. This was followed by a final extension of 72 oC for 10 minutes. The PCR products were separated on a 1% Agarose gel and 1Kb DNA ladder (Fermentas) was used as DNA molecular weight.
Positions of scorable amplified DNA bands were transformed into a binary character matrix (“1” for the presence and “0” for the absence of a band at a particular position). Pairwise distance matrices were compiled by the Numerical Taxonomy System (NTSYS) 2.0 software [16] using the Jaccard coefficient of similarity [17]. Cluster dendrogram was created by unweighted pair-group method arithmetic (UPGMA) cluster analysis [18].
Agarose gel (1%) electrophoresis of PCR products generated after amplification of DNA extracted from each orange fruit following hot water treatment is shown in Figure 1 while genetic analysis using dendrogram revealed three major groups (Group1a, Group1b, and Group 2) of levels of heat resistance among the treated fruits (Figure 2). Group 2 genotype was made up of orange fruits (2, 3, 5, 6, 7, 8, 10, C) with high resistant to heat and orange fruit C (untreated control) was classified into this group because its DNA was never exposed to any heat (Figure 2). This showed that agarose gel electrophoresis of PCR products generated after amplification of DNA extracted from hot water –treated fruits showed that high molecular weight (HMW) bands of DNA were detected successfully for control and fruits treated at 35 oC for 25 and 30 minutes each, 48 oC for 15 and 20 minutes each, 45 oC for 20 and 25 minutes each, 50 oC for 20 minutes and the control (Figure 2).
Meanwhile, group1b genotype was typical of orange fruit (9) with moderate resistant to heat (Figure 2) which implied that HMW bands became fainter for the treated fruits at 50 oC for 15 minutes as a result of temperature –time combination increased (Figure 2). However, Group 1a genotype constituted orange fruits (1, 4) susceptible to heat which implied that HMW bands of DNA were not detected for orange fruits treated at 55 oC for 5 minutes and 75 oC for 5 minutes (Figure 2). The prerequisite for DNA – based analysis is the availability of high quality genomic DNA.
This study has demonstrated that amplification of the RAPD target sequence resulted in one high molecular weight band of the DNA fragment for all the treated fruits. However, the band became fainter as treatment temperature – time increased, thus confirming that the quality of the extracted DNA was affected by the hot water treatments. This was buttressed by the work of [19] who reported significant differences in the DNA quality properties of strawberries during varying hot air treatment – time.
Also, no PCR products were obtained from orange fruits at high temperatures of ≥ 55 oC, confirming that the quality of their DNAs were not detectable for amplification which showed total degradation of DNA in the treated fruits because of the high heat and perhaps this could explained why amber sweet orange fruits treated at temperatures ≥55 oC in the preliminary investigation by [20], deteriorated so rapidly in storage while fruits treated at lower temperatures (≤50 oC), possibly because of high molecular weight (HMW) bands of DNA after treatment had extended shelf life. More so that decay of fruit is the main reason that leads to the degradation of plant genomics. The rapid deterioration could not but also be supported with the earlier observation of [19] that high heat had a negative effect on quality and yield of genomic HMW DNAs extracted from strawberry fruits.
In this study, electrophoresis and PCR analysis showed a progressive reduction of DNA quality during treatments. As temperature-time combination increased, DNA quality decreased significantly. However, hot water treatments at 35 oC for 25 and 30 minutes, 48 oC for 15 and 20 minutes, 45 oC for 20 and 25 minutes and 50 oC for 15 minutes that showed high molecular weight (HMW) bands of DNA of the treated fruits could be used in extending the shelf life of amber sweet orange fruits.
Both authors were actively involved in the research concept and design, collection of data and data analysis, paper writing and final approval of the article.
Authors are grateful to Medical Research Council, Yaba, Nigeria for the laboratory facilities provided to conduct the molecular work.
| Figure 1: Agarose Gel (1%) Electrophoresis of PCR products generated after amplification of DNA extracted from orange fruits treated with hot water
M = 1KB DNA molecular weight marker
-ve = Negative control 1 = 75 oC for 5 minutes 2 = 35 oC for 30 minutes 3 = 35 oC for 25 minutes 4 = 55 oC for 5 minutes 5 = 48 oC for 15 minutes 6 = 48 oC for 20 minutes 7 = 45 oC for 20 minutes 8 = 45 oC for 25 minutes 9 = 50 oC for 15 minutes 10 = 50 oC for 20 minutes C = Control |
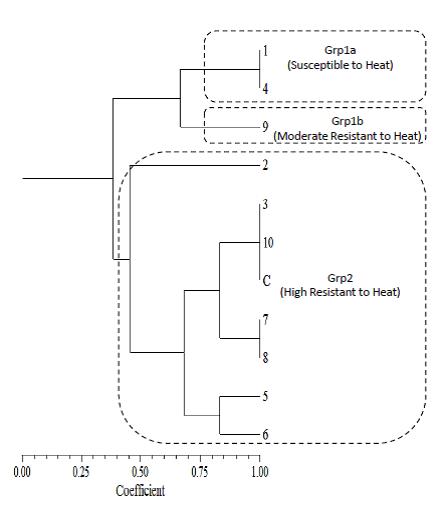 |
| Figure 2: Dendrogram showing the orange fruits levels of genetic resistance to heat
1 = 75 oC for 5 minutes 2 = 35 oC for 30 minutes 3 = 35 oC for 25 minutes 4 = 55 oC for 5 minutes 5 = 48 oC for 15 minutes 6 = 48 oC for 20 minutes 7 = 45 oC for 20 minutes 8 = 45 oC for 25 minutes 9 = 50 oC for 15 minutes 10 = 50 oC for 20 minutes C = Control |






































