Top Links
Journal of Pharmaceutics & Drug Development
ISSN: 2348-9782
How the Artificial Intelligence Tool Psumo-CD is working for Predicting Sumoylation Sites in Proteins
Copyright: © 2020 Chou KC. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
In 2016 a very powerful AI (artificial intelligence) tool has been established for predicting sumoylation sites in proteins, one of the most important post modifications in proteins [1].
Step 1: Opening the web-server at http://www.jci-bioinfo.cn/pSumo-CD, you will see the top page of pSumo-CD on your computer screen, as shown in (Figure 1). Click on the Read Me button to see a brief introduction about this predictor.
Step 2: Either type or copy/paste your query protein sequences into the input box at the center of (Figure 1). The input sequences should be in the FASTA format. For the examples of sequences in FASTA format, click the Example button right above the input box.
Step 3: Click on the Submit button to see the predicted result. For example, if you use the Sequences in the Example window as the input, after a few seconds, you will see the corresponding predicted results, which is fully consistent with experiment observations.
Step 4: Click the Data button to download the benchmark dataset used in this study
Step 5: Click the Citation button to find the relevant papers that document the detailed development and algorithm for iSuc-PseOpt
It is anticipated that the Web-Server will be very useful because the vast majority of biological scientists can easily get their desired results without the need to go through the complicated equations in [1] that were presented just for the integrity in developing the predictor.
Also, note that the web-server predictor has been developed by strictly observing the guidelines of “Chou’s 5-steps rule” and hence have the following notable merits (see, e.g., [2,3] and three comprehensive review papers [4-6]): (1) crystal clear in logic development, (2) completely transparent in operation, (3) easily to repeat the reported results by other investigators, (4) with high potential in stimulating other sequence-analyzing methods, and (5) very convenient to be used by the majority of experimental scientists.
It has not escaped our notice that during the development of iSuc-PseOpt web-server, the approach of general pseudo amino acid components [7] or PseAAC [8] had been utilized and hence its accuracy would be much higher than its counterparts, as concurred by many investigators (see, e.g., [9,10]).
For the marvelous and awesome roles of the “5-steps rule” in driving proteome, genome analyses and drug development, see a series of recent papers [11-32] where the rule and its wide applications have been very impressively presented from various aspects or at different angles.
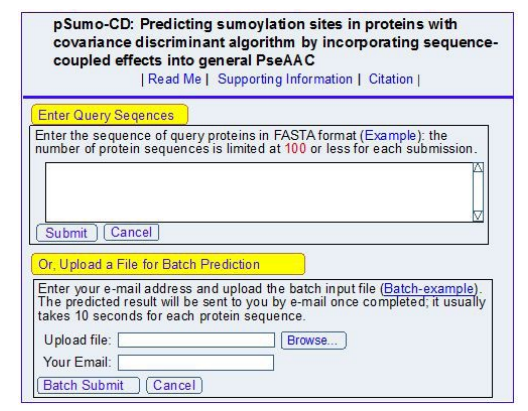 |
| Figure 1: A semi-screenshot of the top-page for the web-server pSumo-CD at http://www.jci-bioinfo.cn/pSumo-CD. (Adapted from [1] with permission) |






































