Top Links
Journal of Health Science Studies
ISSN: 2767-9136
Emerging Coxsackievirus A6 Causing Hand-Foot-and-Mouth Disease in Children in Gabon
Copyright: © 2019 Lekana-Douki SE. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
Hand-Foot-and-Mouth Disease (HFMD) is an epidemic childhood illness caused by enteroviruses including enterovirus A71 (EVA71), coxsackievirus A16 (CV-A16) and coxsackievirus A6 (CV-A6). This disease mainly affects children under 5 years of age, causing typical skin rash such as papulovesicular rash on the palms, feet, and mouth.
Keywords: Hand-Foot-and-Mouth Disease; Coxsachievirus A6; Children; First Description in Gabon
Hand-Foot-and-Mouth Disease (HFMD) is a common epidemic childhood illness caused by viral pathogens within the order Picornaviridales, in the family Picornaviridae, in the genus Enterovirus and the species A. The most common viruses causing outbreaks of HFMD are enterovirus A71 (EV-A71), coxsackievirus A16 (CV-A16) and coxsackievirus A6 (CV-A6) which has emerged in Finland and then worldwide since 2008 [1-5]. The characteristics of this disease defined by the World Health Organization are fever > 38oC in children < 5 years of age, typical skin rash (papulovesicular rash on the palms or soles of the feet, or both, buttocks, knees, or elbows) with or without mouth ulcers [6]. This disease is transmitted by sneezing, by direct or indirect contact with saliva, secretions or feces. Usually, this contagious disease is mild and self-limiting in one week. However, atypical clinical signs may include eruption in others anatomic sites [6]. Severe complications including encephalitis, myocarditis or meningitis have been described, less frequently in association with HFMD [7]. In Europe, a study reported an outbreak of HFMD in 2014 and 2015 in France, during which the most frequent serotype was CV-A6. Moreover, Coxsackievirus A6 infection was sometimes responsible for more than half the atypical form [4,8]. This infection can be benign and completely resolved in about ten days. However, studies described palmar and plantar desquamation and onychomadesis several weeks after the disease [7,9-11]. Furthermore, neurological and systemic complications such as encephalitis can occur after an EV-A71 infection [7,12]. Typical and atypical forms and the severity of the disease could be correlated with the enterovirus serotype [4]. An enterovirus EVA71 vaccine is available in order to prevent severe HFMD [13,14]. In several countries such as China surveillance was established to evaluate the prevalence of this illness and the efficacy of the vaccine against EV-A71. It showed the necessity to develop a multivalent HFMD vaccine including the CV-A16 and emerging CV-A6 pathogen [2,13,14].
Few data on epidemics or cases of HFMD in Central Africa are available. In Gabon this disease associated with enteroviruses is undocumented. Four cases of HFMD from neighborhood children occurred in Franceville in 2018. It is the first clinical description of HFMD and the first molecular characterization of Coxsachievirus A6.
Throat and nasal flocked swabs “553C” (Copan, Diagnostic) were collected, placed in dry tubes and diagnosed for enteroviruses at the CIRMF. All parents gave oral informed consent for the tests. The CIRMF is authorized to establish an epidemiological surveillance and a molecular diagnosis of the diseases circulating in Gabon. Ribonucleic acid (RNA) was extracted from swabs with the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instruction after pretreatment in saline (0.9%). The elution volume of RNA was 100µl.
A Real Time Polymerase Chain Reaction (PCR) was performed using primers for amplification of 5’untranslated region (UTR) of all enteroviruses with the kit SuperScript III One-Step RT-PCR Platinium Taq Invitrogen [15]. Each 25µl reaction mixture contained 5µl of 5X One Step RT-PCR buffer containing 12.5 mM magnesium chloride, 400 µM each deoxynucleoside triphosphate, 40 ng of bovine serum albumin per µl, 0.4 µM primers, 0.2 µM probe and 5 µl of eluted RNA. The PCR was performed using the forward primer 5’-ACATGGTGTGAAGAGTCTATTGAGCT-3’, the reverse primer 5’-CCAAAGTAGTCGGTTCCGC-3’ and the probe 5’-ATTAGCCGCATTCAGGGGCCGGA-3’ labeled at the 5’ends with the FAM quencher and at the 3’ ends with the Black Hole Quencher 1. For genotypic identification a conventional nested PCR was performed using outer forward primer (224-VP3), outer reverse primer (222-VP1), inner forward primer (AN89-VP1) and inner reverse primer (AN88-VP1) targeted at the 357 pb VP1 partial sequence gene [16]. The Kit SuperScript III One-Step RT-PCR Platimium Taq (Invitrogen) was used for the first round. Each 25-µl reaction mixture contained 1X buffer, 0.1 mM of magnesium sulfate, 40 ng/µl of bovine serum albumin, 0.4 µM of primers, 1 µl of Taq and 5µl of eluted RNA. The Kit DNA+Taq Polymerase (Invitrogen) was used for the second round with the same final volume and concentration of reagent, 1 µl of Platinium Taq (Invitrogen) and 2µl of amplification product of the first PCR. The first round was done with the following conditions: 30 min at 42 o C, 3 sec at 95 o C, followed by 10 cycles at 95 o C for 20 sec, a decrease of one degree per cycle from 52 o C to 43 o C and 72 o C for 1 min, followed by 40 cycles at 95 o C for 20 sec, 42 o C for 30 sec and 72 o C for 1 min, and a final elongation at 72 o C for 10 min. The second round of PCR was run for 3 min at 95 o C, followed by 45 cycles at 95 o C for 15 sec, 42 o C for 20 sec and 72 o C for 30 sec, followed by 72 o C for 10 min.
A second conventional PCR was performed using two rounds with the same kit and concentration of reagent as previously and specifics nested primers based on the CV-A6 Gdula strain of VP1 to determine the genotype (outer sense 5’-GARGCTAACATYATAGCTCTTGGAGC-3’, outer antisense 5’-CCYTCATARTCHGTGGTGGTTATGCT-3’, inner sense 5’-GACACYGAYGARATYCAACAAACAGC-3’, inner antisense 5’-CGRTCRGTTGCAGTGTTWGTTATTGT-3’ [3]. The first round was done with the following conditions: 30 min at 50 o C, 10 min at 95 o C, followed by 40 cycles at 94 o C for 40 sec, 53 o C for 40 sec and 72 o C for 1 min, and a final elongation at 72 o C for 10 min. The second round of PCR was run for 10 min at 95 o C, followed by 40 cycles at 94 o C for 40 sec, 53 o C for 40 sec and 72 o C for 1 min, and a final elongation at 72 o C for 10 min. These generated amplicons of 891 pb were purified and sequenced by the method of Sanger with a 3500 Genetic Analyzer (Applied Biosystems) using a BigDye Terminator Ready Reaction Cycle sequencing kit. Phylogenetic analyses were performed using a multiple sequence alignment of the sequence obtained and a selection of reference strains from the GenBank database using the Basic Local Alignment Search Tool (BLASTn). Phylogenetic relationship was determined with MEGA 4.0 software. Multiple sequence alignments were created using ClustalX (version 1.81). The phylogenetic tree was built using the maximum-likelihood method with the PhyML algorithm [17] and drawn using FigTree v.1.4.0.
In March 2018 three children aged three years old, from the same neighborhood and school, and, a sibling aged 13 months visited the medical office of the Centre International de Recherches Médicales de Franceville (CIRMF) showing symptoms of hand-foot and-mouth disease (Table 1). All four children had a rash on the palms, mouth, feet and knees and fever (38oC). The infant had an additional atypical form and showed a vesicle rash on the upper limbs and shoulders. One of three-years-old children who had an additional eruption on the elbows and a runny nose presented a palmar and plantar desquamation and onychomadesis one month after the symptoms disappeared (Table 1).
Enteroviruses were detected in the four samples. The child with a palmar and plantar desquamation and onychomadesis had the highest viral load. Four identical sequences were obtained from the samples of the four children. The sequence (807 pb) from the four isolates, named GAB01CoxA is available in the DDBJ/EMBL/GenBank database under accession numbers MK433298. A phylogenetic analysis based on 807 nucleotides sequence of the VP1 gene was performed using the sequence GAB01CoxA, 41 sequences of Coxsackievirus A6, from the GenBank database (Figure 1). We used the sequence of the Gdula strain prototype (genotype A) isolated in the USA in 1949 and genotype sequences B, C and D. The Gabonese strain displayed 99% to 94% identity at the nucleotide level with GenBank strains of genotypes D4 circulating in Europe and Asia between 2014 and 2017 (Figure 1). The Catalonia strain circulated in Spain in 2017 (Accession no. MK167082) have 99% identity with the Gabonese strain.
We reported four cases of HFMD among neighbor children under five years old. These cases occurred in Franceville, a town in the south of Gabon, in March 2018. Molecular diagnostics showed that an enterovirus infected the patients. More specific molecular investigations showed that the children were infected by Coxsackievirus A6. These results corroborated those of several studies in Europe and Asia that reported the circulation of Coxsackievirus A6 causing HFMD [2,4]. One of the children showed an atypical form with a vesicle rash on the upper limbs and shoulders. Another had palmar and plantar desquamation and onychomadesis one month after contracting HFDM. These two atypical dermatologic presentations correlate studies which reported that CV-A6 was more frequently associated with unusual forms compared to other enterovirus serotypes [4,7,18]. However, the association between CV-A6 infection and onychomadesis is still widely debated in the literature [7]. Alignment was made with the four sequences (807 pb) obtained showing that they were strictly identical. The strain of this report (isolate GAB01CoxA, accession number MK433298) clustered with Coxsachievirus A6 and belonged to subgenotype D4. The homology (99%) between the Gabonese strain and this Spanish strain suggests that the Gabonese strain was recently imported in Gabon. However, the low number of cases and the lack of data concerning the circulation of CVA6 in Gabon, does not exclude their circulation and does not allow us to conclude that this strain is endemic or imported. An epidemiological survey was carried out. Patients did not travel outside Gabon during two months before onset. However, there are many exchanges and travels between Franceville, Europe and China for tourist and especially professional reasons. Since 2010, it would seem that most of the outbreaks of CV-A6 worldwide were due to genotype D3 [19]. However, these past few years, subgenotypes D4 and D5 emerged in Europe and Asia [3]. Subgenotype D4 was described in Finland and Germany, in 2008 and 2014 respectively. In China, several subgenotypes including D4 circulated in 2010, 2013 and 2015. Despite the recent emergence of D4, publications described a lower circulation [3,20]. In Central Africa and Gabon, little data on HFDM were available. No studies mentioned the circulation of CV-A6 and the genotypes circling in Gabon. It may be due to the fact that this disease has not been described much because the clinical symptoms resemble those of a mild allergy. Indeed, other children from the same school and city appeared to have had the disease in the previous weeks even though no diagnosis was made. Thus, the health centers of the city were informed in order to set up a surveillance network of this disease. The number of children affected by HFDM in the same time period could have been underestimated. In France, it was showed that there was a gap existed in knowledge of the epidemiology and clinical impact of HFMD or herpangina because virological diagnosis wasn’t performed. Therefore, the National Reference Laboratory for Enterovirus and Parechovirus set up surveillance in different regions of France from April 2014 to March 2015 reported that CV-A6 dominated the two epidemic occurred during this period [4]. These four cases of HFMD, described for the first time in Gabon suggested that CV-A6 circulated in the country and probably that ambulatory pediatrics cases could not have been investigated for virological diagnosis such as reported in France. A surveillance for HFMD would allow a better understand of the burden of HFMD, the epidemiology of enteroviruses such as CV-A6 and their serotypes. Previous studies have described that enteroviruses involved in HFMD can induce neurological complications [7,12,20]. Furthermore, a surveillance network of neurological syndrome has already been established in Franceville and Libreville, the capital of Gabon. So, first, we will include the diagnosis of Coxsackievirus in the surveillance system of neurological syndrome. Simultaneously, we set up a surveillance system of HFMD in the two main hospitals of Franceville. We informed the medical staff by describing the WHO case definition and the samples will sent to the CIRMF with clinical and epidemiological data for viral diagnosis.
These preliminary data which highlights the circulation of Coxsackievirus A6 in children under five years old emphasize the importance of establishing surveillance for HFMD in Gabon. We started the establishment of this network including also the viral diagnosis of neurological disorders.
We acknowledge Heïdi Lançon for the English revision of the manuscript. We also wish to thank Kassa Kassa Roland Fabrice doctor at the CIRMF.
The authors declare no conflicts of interest.
The “Centre International de Recherches Médicales de Franceville” (CIRMF) is authorized to establish an epidemiological surveillance and a molecular diagnosis of the diseases circulating in Gabon. All parents of the four children gave verbal consent. The work described has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki).
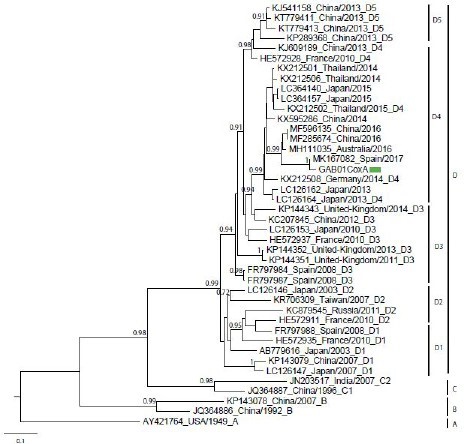 |
| Figure 1: Molecular characterization of coxsackievirus strains; Phylogenetic tree for nucleotide sequences of Coxsackievirus A6 strains VP1 gene. The green rectangle indicates the sequence of Gabon (GAB01CoxA) |
|
Case 1 |
Case 2 |
Case 3 |
Case 4 |
Age |
3-years-old |
3-years-old |
13-months-old |
3-years-old |
Sex |
Male |
Male |
Male |
Male |
Week of the onset symptom |
Week 11 (2018) |
Week 11 (2018) |
Week 11 (2018) |
Week 12 (2018) |
Epidemiological linkage |
Same school and same class as cases 2 and 4 |
Same school and same class as cases 1 and 4 |
Brother of the case 2 |
Same school and same class as cases 1 and 2 |
Fever |
38 oC |
38 oC |
38.8 oC |
38 oC |
Cutaneous symptoms |
Vesicular skin rash on mouth, hands, |
Vesicular skin rash on mouth and feet |
Vesicular skin rash on mouth, hands, arms, shoulders, knees and feet |
Vesicular skin on mouth, hands and feet |
Others symptoms |
Palmar and plantar desquamation and onychomadesis after one month |
No other symptoms |
No other symptoms |
Runny nose |






































