Top Links
Journal of Antibiotics Research
ISSN: 2574-5980
Genetic Relatedness and Characterization of O25b-B2-ST131, in Stool Isolates of Extended-Spectrum Cephalosporin-Resistant Escherichia Coli Strains in Healthy Children under 10 Years of Age
Copyright: © 2023 Ashraf Mohabati Mobarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Related article at Pubmed, Google Scholar
In the pre-anesthesia era, the anguish of planned surgical maneuver was dreadful and the experience of actual procedure “utterly speechless torture”. Although, the concept of pain relief and even total insensibility was not unfamiliar to medical profession, some of the “Big Giants in Medicine” believed “knife and pain as inseparable” and the efforts to relieve or prevent pain “in vain”. In such a situation the anesthesia,one of the greatest boons of science for mankind, “burst like a revolution on medical profession”. Despite hostility from religious sect, professional colleagues and civil societies the pioneers in discovery of anesthesia stood firm, unshaken by the negative criticism. Although the fate of the active contestant of “Ether Controversy” was mournful,their untiring and dedicated struggle to relieve the sufferings of mankind can never be underscored. Renowned Arab surgeon Ibn al Quff (1232-1286 AD) was the first to suggest anesthesia as independent speciality. However, it took almost 700 years for his dream to come true.
Background and Purpose: Escherichia coli (E. coli) is one of the multidrug-resistant pathogens, producing extendedspectrum beta-lactamase enzymes. Molecular typing of this pathogen can be useful for determining the source of dissemination and transfer of resistance and virulence genes of these isolates. Considering the significance of infection in children, in this study, we examined the stool flora of children (< 10 years) to show the characteristics and clonal relationship of the isolates.
Materials and Methods: In this research, we used 100 isolates of E. coli, resistant to third-generation cephalosporins and extended-spectrum beta-lactamase (ESBL) genes, isolated from healthy (non-diarrheal) children under 10 years of age. Phylotyping (B2 and D), serogrouping (O25, O16), plasmid replicon typing, O25b-B2-ST131 clonal group and pulse field gel electrophoresis was performed. The data were analyzed using SPSS software, version 25.
Results: Twenty-seven percent of samples belonged to phylogroup B2 and 16% to phylogroup D/E. PCR identified 18 antibiotic resistance plasmid genes. Two isolates belonged to B2-phylogroup and serogroup O25 and one had mutations in two genes, identified as clonal group O25b-B2-ST131. Among diverse bands (11-20 bands; average 17 with a size of 20-1135 kb), 19 pulsotypes were obtained, four had more than one isolate, and formed a cluster. Several common genes were identified among isolates.
Conclusions: The presence of E. coli phylogroup B2 in healthy children under 10 years of age with a very diverse genotype in our study showed this pathogen an important reservoir of virulence and resistance genes. As these strains can become pathogenic, it is important to pay greater attention to prevent the spread of these isolates in schools, hospitals, and communities.
List of Abbreviations: HE: Red Crescent Nursery, PU: Pooyan Nursery, CH: Chamran Hospital, F: Ashna family, ALI: Ali Asghar Hospital, FA: Farkhondeh Hospital, BE: Besat Rehabilitation, AM: Amenah Rehabilitation, A: Andarzogo Rehabilitation, KH: Khayam Rehabilitation, NA: Narmak Rehabilitation, MO: Morteza Gard Rehabilitation
Escherichia coli (E. coli) is one of the first bacterial species that is colonized in the intestines of babies, and it can usually be detected in the baby's feces a few days after birth [1]. Environmental studies have shown that commensal strains have acquired or lost certain traits such as antibiotic resistance, virulence, serological changes, and even biochemical characteristics during their dissemination [2]. One of the problems related to these commensal strains is acquiring antibiotic-resistant genes, carrying plasmids, and spreading them. E. coli resistant to third-generation cephalosporins are a great threat not only in hospitals but also in the community [3]. It is a concern that antimicrobial resistance in commensal bacteria can be transferred to pathogenic bacteria, mainly by plasmids [4]. Genes producing extended-spectrum beta-lactamase (ESBL) enzymes are often encoded on transferable plasmids that encode resistance genes, and the acquisition of these resistance genes by commensal or fecal isolates, in turn, leads to multidrug-resistant (MDR) pathogens [5]. In recent years, extraintestinal infections caused by these MDR strains such as ST131 are increasing rapidly [6]. ST131 is a global epidemic clonal group and MDR clone. The emergence of high rates of antimicrobial resistance in E. coli O25b-B2-ST131 is a crisis with limited treatment options to eradicate the infection and increased morbidity and mortality rates [7]. E. coli is one of the most common ESBL bacteria, and certain phylogroups such as phylogroup B2, and its clone ST131 are associated with global distribution [8]. Phylogroup B2 has a high staying power in the intestines and plays a major role in extraintestinal infections (ExPEC) [9]. Therefore, finding these phylogroups in healthy carriage children can be significant and is considered a dangerous warning factor for further clinical infections. the use of molecular typing techniques, including PFGE (pulsed-field gel electrophoresis) and plasmid replicon typing, can be useful for determining the source of dissemination and transfer of resistance and virulence genes of these isolates [10]. Using PFGE (Pulse-field gel electrophoresis) as a gold standard typing method to understand clonal relatedness between isolates and comparing data, including phylogroup types and resistance plasmids, can help the healthcare system track the sources of infection and the distribution of potential resistance genes.
So far, there have been few reports about the stool flora of healthy children in Iran [11], and most of the studies focused on clinical samples. Therefore, examining the stool flora of these children and showing the isolates’ clonal relationship can be significant in controlling the infection.
This descriptive-research study was conducted from January 2020 to May 2022 at Tarbiat Modares University, Tehran, Iran. In this research, we used 100 isolates of E. coli resistant to third-generation cephalosporins (CTX, CAZ=100%) and ESBL genes (TEM=26%, CTX-M1=98%, SHV=51%), which were isolated from all healthy children under 10 years of age (non-diarrheal) and were kept in the archives of the bacteriology department. of these, 57% were boys and 43% were girls. Ethics approval for this study was obtained from the Ethics Committee of the Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran (IR.MODARES.REC.1399.085).
To confirm these 100 E. coli isolates, various biochemical tests were performed. Each isolate was first cultured on EMB (Eosin Methylene Blue) Agar. After 24 hours, when the metallic green color was observed, it was inoculated in TSI (Triple sugar Iron) Agar and SIM (Sulfide Indole Motility) medium, respectively, in terms of glucose and lactose sugar fermentation, as well as the production of indole, hydrogen sulfide, and movement. By culturing bacteria in Simon's Citrate medium, the isolates were evaluated for citrate consumption. Moreover, all isolates were tested for MR-VP (Methyl Red, Vege-Prosquare) reaction. Urea broth medium was also used to investigate urease enzyme production. After the biochemical confirmation of each isolate as E. coli, the bacteria were stored in 300 µl of Trypticase medium (Merck-Germany) with 15% glycerol at minus 70°C.
Fresh overnight cultures were prepared, and DNA extraction was done using the boiling method [12].
The O25, O16 serogroups [13], phylogenetic group distribution of the E. coli isolates (B2, D) was specified using the Clermont quadruplex polymerase chain reaction (PCR) method, which detects the presence or absence of four DNA markers (yjaA,TspE4.C2, chuA and arpA) [14] (table 1).
The multiplex PCR reaction for replicon typing plasmid was done using the primers and the E. coli DH5α isolate was used as a negative control [15] (table 1).
To identify ST131 clonal group, all isolates of phylogroup B2 and serotype O25, D phylogroup, and O16 serogroup were screened using a single nucleotide polymorphism PCR in the protected genes mdh36 and gyrB47, related to ST131. In case of difference in mdh (C525T, C288T) and gyrB47 (T735C, C729T, C621T) fragments, the bacterium belonged to another clonal group [16] (table 1)
In order to analyze the clonal relationship of genomic DNA, according to the PulseNet One-Day (24–28 hr) Standardized Laboratory Protocol, 27 isolates belonging to phylogroup B2 were subjected to PFGE (Pulse field gel electrophoresis), and Salmonella enterica serotype Braenderup strain H9812 was used as a reference marker. Also, XbaI endonuclease enzyme (Thermo Scientific, Waltham, MA) was used to digest the genomic bands. It was isolated using a CHEF Mapper (II) system (Bio-Rad Laboratories) using 1% LFTM agarose (X174; Amresco, Solon, OH). The photos were analyzed in TIFF (Tagged Image File Format) using Gel-Compar II V.5.1 (Applied Maths, Belgium), and the cluster analysis of the Dice similarity coefficient through UPGMA (unweighted pair group method with arithmetic means, band tolerance: 1.5%). The distance matrix between subtypes was also done for clustering. The isolates that had a cut-off ≥ 95% of their band patterns were considered to belong to the same clonal lineage (cluster).
The data were analyzed and compared using SPSS software (version 25; Inc., Chicago, IL, USA). The analysis of variance and unpaired t test were used. A confidence interval (CI) of 95% was considered, and P< 0.05 was considered statistically significant.
Phylogrouping (B2, D), done according to the Claremont method, showed that 27% of the samples (27/100 isolates) belonged to phylogroup B2 and 16% belonged to phylogroup D/E (16/100 isolates). To separate phylogroups D from E, the MLST test should have been used to identify the isolates belonging to the ST131 clone; therefore, we could not include these phylogroups in the study.
By using PCR technique and specific primers, 18 antibiotic resistance plasmid genes (HI2, KIB, HI1, FreP, I1-γ, X, L/M, N, FIA, FIB, FIC, W, Y, P, A/C, T, K, B/O) were identified; 55% of the isolates (55/100 isolates) had FreP gene, 19% had I1 plasmid, and 13% (13/100 isolates) had FIB plasmid. K/B and Y, each with 5% (5/100 isolates), and B/O and FIA, each with 4% (4/100 isolates), were also identified among the isolates. Three isolates with P plasmid and A/C, HI2, and FIIA plasmids were identified, each in one isolate. In addition, FIC, L/M, W, X, N, T, and HI1 plasmids were not found among the isolates.
Two isolates belonged to B2-phylogroup and serogroup O25 (isolates no. 299 and 120); only isolate no. 120 had mutations in two genes gyrB47 and mdh36, which belonged to a 7-year-old boy, identified as clonal group O25b-B2-ST131. Among the isolates, we did not identify phylogroup D.
In this study, diverse bands were obtained and, in all samples, they showed 11 to 20 bands (on average 17 bands) with a size of 20-1135 kb. Considering 95% similarity, 19 pulsotypes were obtained from all isolates, among which four pulsotypes included more than one isolate and formed a cluster, and 15 single clones were obtained. The largest pulse type included five isolates with a completely similar pattern (figures 1 and 2).
In the drawn dendrogram (figure 2), the information related to children’s age and sex, phylogroup B2, subgroup B2, serogrouping, virulence factors, plasmid replicons, resistance genes, and isolation location of the isolates have been investigated. Five isolates;No. 174, 175, 180, 185, and 186(cluster I), isolated from a 4-year-old girl, 9-year-old girl, 4-year-old girl, 5-year-old girl, and a 1- year-old boy, respectively, all from the same center (Farkhandeh Hospital), had common malX virulence gene. In addition, isolate 175 had papGIII gene and isolate 186 had papGIII and usp genes. FreP, I1, FIB, FIA, and Y replicons were observed in these isolates, and CTX-M1 was the common resistance gene in these five isolates, three isolates belonging to subtype V phylogroup B2 (STc144), and one isolate (No. 174) belonging to subtype I (STc131). Three isolates (No. 174, 175, and 180) had unknown serogroups, one isolate (No. 185) belonged to serogroup O18 and one isolate (No. 186) belonged to serogroup O75. Pulsotype 181 had 97% similarity with these five isolates, and its isolation location is also the same as the other five isolates. This pulsotype was isolated from a 4-year-old girl, had malX, papGIII, and usp virulence genes, FreP, I1 and FIB replicon types, and CTX-M1 in terms of having resistance genes. This clone has an unknown serogroup and belongs to group V (STc144) in terms of phylogroup B2 subtypes.
Another observed cluster included two isolates 64 and 127, which were isolated from a 6-year-old and a 5-year-old girl, respectively, but their isolation location was different (cluster III). Isolate No. 127 had an unknown serogroup and isolate No. 64 had an O2 serogroup. Among these two isolates, isolate No. 127 had usp, malX, and papGII virulence factors, and isolate No. 64 had usp, while Frep plasmid was common in both of them; isolate No. 64 also had FIB. In terms of resistance genes, both of them had CTX-M1, and isolate No. 127 also had TEM. Both of these isolates had subtypes I (STc131), III (STc127), and V (STc144), in terms of placement in phylogroup B2 subtypes, while isolate No. 64 had subtype IX (STc95), as well. Pulsotype 82, isolated from a 4-year-old girl, had 86% similarity with these two isolates (No. 64 and 127) and had CTX-M1 resistance gene, usp, malX, and papGII virulence factors, and FreP type plasmid. In terms of serogrouping, it belonged to serogroup O2, like isolate No. 64, and in terms of subtype, group B2, it had subtypes V (STc144) and I (STc131), like isolate 127.
The other cluster was related to two isolates No. 308 and 309, isolated from the same place, from a 2-year-old boy and a 7-year-old girl, respectively (cluster IV). These two isolates had replicon type I1 and CTX-M1 resistance genes in common and both belonged to subtype IX (STc95). Isolate No. 308 belonged to serogroup O2 and lacked virulence factor, while isolate No. 309 had malX and an unknown serogroup. Isolate No. 131 was also 82% similar to these two isolates (No. 308 and 309), as it had an unknown serogroup, CTX-M1 resistance gene, and similar to isolate 309, it had B2 subtypes V (STc144) and IX (STc95). Isolate No. 131 was isolated from a 6-year-old girl who had FreP plasmid replicon and papGII virulence gene.
In this study, PFGE (Pulse field gel electrophoresis) results indicated high genetic diversity and several isolates with a common source. In some cases, despite the common source of isolates, the same pulsotypes were observed with different virulence factors and different plasmid replicons; and in some other cases, the same virulence factors were observed with the same plasmid replicons and different pulsotypes. In another study on human feces, 15 isolates were found to represent the dominant antibiotic resistance patterns for E. coli populations [17], which is in line with the diversity of E. coli genotypes, found in the present study. Among the reasons for the high diversity of some pulse types in our study (19 pulsotypes), we anticipate that these isolates were possibly not from the same clone or common origin and thus did not have the same genotype. Besides, the variety of colon isolates and genetic exchange by the phenomenon of horizontal gene transfer, number, and size could play a role in this high diversity. It has to be noted that pulse types of different studies cannot be easily compared, because the bands obtained from the different populations around the world vary from each other and patients with different medical conditions [18,19]; also, we believe that the different laboratory conditions, sampling location, and type of samples collected in different parts of the world play a role in this difference. Therefore, we cannot compare the results of our study with other studies.
One of the notable points in genotyping (of these 27 isolates) in our study was that despite being in the same phylogroup (phylogroup B2) and similar drug resistance pattern, serogrouping showed many differences in virulence factors. In contrast to IncF group plasmids, it was the dominant plasmid in these isolates, which shows the high ability of these isolates to acquire and transfer new traits. Our findings are in accordance with other studies conducted (in Iran and the world) on human feces samples, in which it was found that most E. coli isolates mainly belong to the B2 phylogenetic group [20,21]. In our previous study, we also showed that B2 phylogroups had the highest virulence factors and resistance genes [22]. In another study on fecal E. coli (N= 1283 isolates), in addition to showing phylogroup B2 as the most common (38.3%), the researchers also showed that this phylogroup had the highest prevalence of bacteriocinogeny, as well [21]. In addition, the association of E. coli strains, belonging to phylogroups B2 with advanced colorectal neoplasia [23] and resident strains of E. coli (detectable in human intestine for months, compared with transient strains, which persist for days to weeks) [24] have also been noted in other studies. These results indicate the significance of E. coli in medicine.
In the study by Bahadori and colleagues, which aimed to compare phylogenetic groups of fecal microbiota (by PFGE) in patients with urinary tract infection (UTI) and healthy subjects, the results revealed that most ExPEC strains from the urine and feces of healthy women belonged to the phylogenetic group B2, followed by D [25]. In the study of Contreras-Alvarado et al in Mexico., 126 strains from children hospitalized with complicated urinary tract infections (cUTIs) were identified as O25b, which had high genetic diversity. ST131 (63.63%) was mainly identified with phylogenetic group B2. These strains showed ≥80% similarity, indicating a highly related profile [34]. These results are also consistent with the results of our study in terms of B2 phylogroups. Other studies have also determined these two phylogenetic groups (B2 and D) as the most frequent cause of ExPEC infections [26]. According to the findings obtained in our research and similar studies, it can be concluded that phylogroup B2 isolates play a major role in extraintestinal infections and carry resistance genes, which can lead to the spread in hospitals and patients and result in resistant hospital infections. Considering the heterogeneity of some E. coli isolates in the present study and the lack of significant relationship between them in terms of resistance and/or virulence genes, it can be concluded that these isolates did not have a clonal relationship and in general, transmission between individuals is less noticeable in these isolates. However, because of the high similarity percentage of some studied isolates, it is likely that these isolates are of the same origin and have undergone genetic changes over time. As you can see in the drawn dendrogram, most of the isolates with the same genomic pattern were isolated from the same place. We speculate that this finding is related to the center, from which the five dominant isolates of the present study were separated. Since they include about 18% of the isolates, it is suggested that they consider a new solution for maintaining hygiene and preventing the spread of these strains.
The identification of the IncF incompatibility group, in our study, indicates that this incompatibility group plays an important role in the transfer of ESBL coding genes. Among the different types of this group, both IncFI and IncFII are involved in blaCTX-M15 gene transfer. These results showed that children under 10 years of age are very variable in terms of genotype and are the reservoirs of a collection of antimicrobial resistance plasmids. There were similar profiles in some isolates, as well; for example, isolates No. 180, 181, and 193 had the same plasmids as FreP, I1, FIB, the same virulence factor malX, unknown serogroup, phylogroup B2 and subtype V (polB). Most of the ST131 E. coli strains are ESBL-positive and produce CTX-M-1, while TEM and SHV are rarely observed among them, and they are resistant to cephalosporins and fluoroquinolones [27]. In the present study, we also found an isolate of O25-B2-ST131. This isolate was CTX-M-1 positive and lacked TEM and SHV; it was isolated from a 7- year-old boy and had usp and malX genes. In the study by Shokouhi Mostafavi and colleagues on 248 isolates, separated from samples with pyelonephritis, six O25-B2-ST131 isolates were identified; all of which were resistant to ceftazidime, cephazolin, coamoxiclav, and amoxicillin [16]. This finding suggests that an isolate with the same profile was circulating among children. These results are consistent with others in terms of frequency and type of plasmids detected. Based on the results of the present study, O25b:H4-B2-ST131 accounted for 61% of ESBL-positive E. coli; these isolates are often resistant to broad-spectrum cephalosporins, carbapenems and CAZ/AVI can also become resistant and can lead to a crisis in the treatment of related infections [28]. The identification of usp and malX in the present study showed the higher significance of these strains, as these characteristics can result in higher virulence and pathogenicity to persist in human microbiota and make them capable of the development of UTI [29]. In another study on fecal samples of Iranian children, O127 and O128 serotypes were identified as the majority of enterotoxigenic E. coli and more than half of the strains (57%) were resistant to more than one antimicrobial agent [30]. Also, in the study by Huang and colleagues on 157 stool samples from children aged 0 to 18, they found O25b-ST131 positive in 80% of E. coli strains, as the most common ESBL-producing E. coli [31]. Another study on children in Qatar showed aap, capU, and aggR virulence genes with the highest frequency (65, 60, and 55%, respectively) [32]. Although the identified genes vary among studies, all these studies indicate the high virulence and pathogenicity of E. coli strains of the children’s microbiota, which calls for greater attention to this strain. Considering the antibiotic resistance of most isolates in the present study, preventive measures are necessary for the prevention of the spread of E. coli in schools and communities [33].
Our results showed that E. coli phylogroup B2 was present in healthy children under 10 years of age with a very diverse genotype that can be an important reservoir of virulence and resistance genes. As these strains can become pathogenic or become extraintestinal under the right conditions and time or their resistance genes can be transformed into pathogenic pathotypes, it is important to pay greater attention to prevent the spread of these isolates in schools, hospitals, and communities.
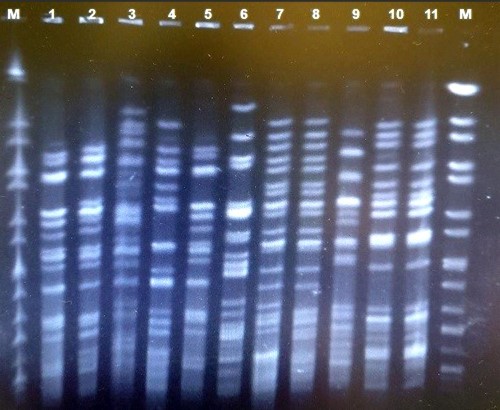 |
| Figure 1: PFGE gel image after enzymatic digestion with XbaI and electrophoresis: M wells: marker of Salmonella serotype related to Branederup strain H9812, wells (1-11) E. coli phylogroup B2 isolates./td> |
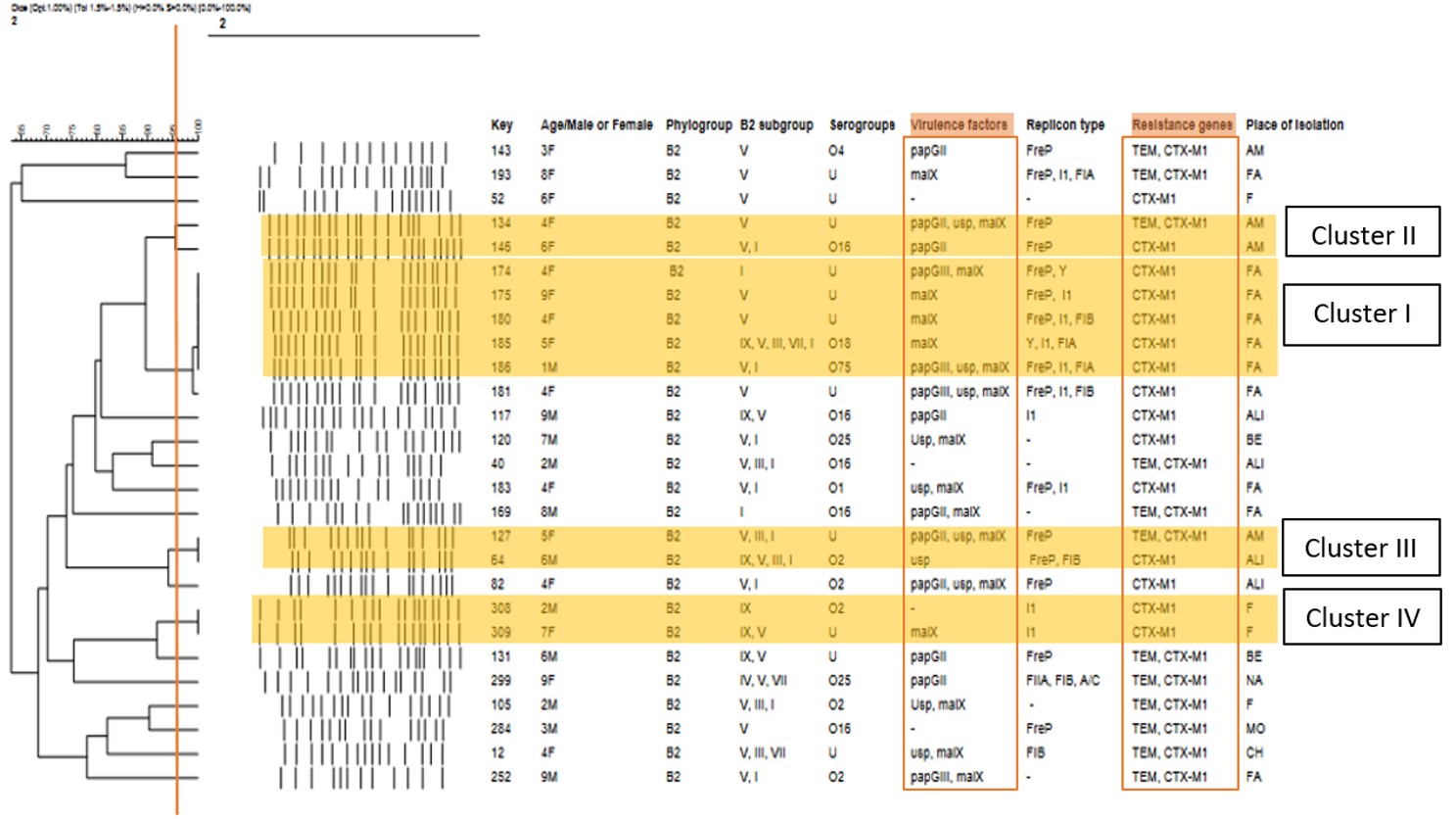 |
| Figure 2: Similarity dendrogram of PFGE profiles of 27 E. coli isolates, belonging to phylogroup B2 |
Target gene |
Primers, Genes, and Sequence (5'- 3') |
Annealing time & temp |
Amplicon size (bp) |
Reference |
chuA |
F: GAC GAA CCA ACG GTC AGG AT |
55°C |
279 |
|
R: TGC CGC CAG TAC CAA AGA CA |
||||
yjaA |
F: TGA AGT GTC AGG AGA CGC TG |
55°C |
211 |
(14) |
R: ATG GAG AAT GCG TTC CTC AAC |
||||
TspE4C2 |
F: GAG TAA TGT CGG GGC ATT CA |
55°C |
152 |
|
R: CGC GCC AAC AAA GTA TTA CG |
||||
arpA |
F: AACGCTATTCGCCAGCTTGC |
56°C |
400 |
|
R: TCTCCCCATACCGTACGCTA |
||||
O25 |
R: GAGATCCAAAAACAGTTTGTG |
59°C |
313 |
(13) |
O16 |
R: GGATCATTTATGCTGGTACG |
59°C |
450 |
|
gndbis. |
F: ATACCGACGACGCCGATCTG |
- |
5000-17000 |
|
B/C |
F: GCGGTCCGGAAAGCCAGAAAAC |
60°C |
159 |
(15) |
R: TCTGCGTTCCGCCAAGTTCGA |
|
|||
FIC |
F: TCTGCGTTCCGCCAAGTTCGA |
60°C |
262 |
|
R: GTGAACTGGCAGATGAGGAAGG |
|
|||
A/C |
F: TTCTCCTCGTCGCCAAACTAGAT |
60°C |
465 |
|
R: GAGAACCAAAGACAAAGACCTGGA |
||||
P |
F: ACGACAAACCTGAATTGCCTCCTT |
60°C |
534 |
|
R: CTATGGCCCTGCAAACGCGCCAGAAA |
||||
T |
F: TCACGCGCCAGGGCGCAGCC |
60°C |
750 |
|
R: TTGGCCTGTTTGTGCCTAAACCAT |
||||
K/B |
F: CGTTGATTACACTTAGCTTTGGAC |
60°C |
160 |
|
R: GCGGTCCGGAAAGCCAGAAAAC |
||||
W |
F: TCTTTCACGAGCCCGCCAAA |
60°C |
242 |
|
R: CCTAAGAACAACAAAGCCCCCG |
||||
FIIA |
F: GGTGCGCGGCATAGAACCGT |
60°C |
270 |
|
R: CTGTCGTAAGCTGATGGC |
||||
FIA |
F: CTCTGCCACAAACTTCAGC |
60°C |
462 |
|
R: CCATGCTGGTTCTAGAGAAGGTG |
||||
FIB |
F: GTATATCCTTACTGGCTTCCGCAG |
60°C |
702 |
|
R: GGAGTTCTGACACACGATTTTCTG |
||||
Y |
F: CTCCCGTCGCTTCAGGGCATT |
60°C |
765 |
|
R: AATTCAAACAACACTGTGCAGCCTG |
||||
I1 |
F: GCGAGAATGGACGATTACAAAACTTT |
60°C |
139 |
|
R: CGAAAGCCGGACGGCAGAA |
||||
FreP |
F: TCGTCGTTCCGCCAAGTTCGT |
60°C |
270 |
|
R: TGATCGTTTAAGGAATTTTG |
||||
X |
F: GAAGATCAGTCACACCATCC |
60°C |
376 |
|
R: AACCTTAGAGGCTATTTAAGTTGCTGAT |
||||
HI1 |
F: TGAGAGTCAATTTTTATCTCATGTTTTAGC |
60°C |
471 |
|
R: GGAGCGATGGATTACTTCAGTAC |
||||
N |
F: TGCCGTTTCACCTCGTGAGTA |
60°C |
559 |
|
R: GTCTAACGAGCTTACCGAAG |
||||
HI2 |
F: GTTTCAACTCTGCCAAGTTC |
60°C |
644 |
|
R: TTTCTCCTGAGTCACCTGTTAACAC |
||||
L/M |
F: GGCTCACTACCGTTGTCATCCT |
60°C |
785 |
|
R: GGATGAAAACTATCAGCATCTGAAG |
||||
mdh36 |
F: CTGCAGGGGCGATTCTTTAGG |
65°C |
274 |
(16) |
R: GTTTAACGTTAACGCCGGT |
||||
gyrB47 |
F: GGTAACACCAGAGTGACCA |
60°C |
132 |
|
R: CGCGATAAGCGCGAC |
Sr. No |
Target gene |
Primers, Genes and Sequence (5'- 3') |
AmpliconSize (bp) |
Annealing time & temp |
Reference |
|
1. |
chuA |
F |
GAC GAA CCA ACG GTC AGG AT |
279 |
55°C |
(14) |
R |
TGC CGC CAG TAC CAA AGA CA |
|||||
2. |
yjaA |
F |
TGA AGT GTC AGG AGA CGC TG |
211 |
55°C |
|
R |
ATG GAG AAT GCG TTC CTC AAC |
|||||
3. |
TspE4C2 |
F |
GAG TAA TGT CGG GGC ATT CA |
152 |
55°C |
|
R |
CGC GCC AAC AAA GTA TTA CG |
|||||
4. |
arpA |
F |
AACGCTATTCGCCAGCTTGC |
400 |
56°C |
|
R |
TCTCCCCATACCGTACGCTA |
|||||
5. |
O25 |
R |
GAGATCCAAAAACAGTTTGTG |
313 |
59°C |
(13) |
6. |
O16 |
R |
GGATCATTTATGCTGGTACG |
450 |
||
7. |
gndbis |
F |
ATACCGACGACGCCGATCTG |
5000-17000 |
- |
|
8. |
B/C |
F |
GCGGTCCGGAAAGCCAGAAAAC |
159 |
60°C |
(15) |
R |
TCTGCGTTCCGCCAAGTTCGA |
|||||
9. |
FIC |
F |
GTGAACTGGCAGATGAGGAAGG |
262 |
60°C |
|
R |
TTCTCCTCGTCGCCAAACTAGAT |
|||||
10. |
A/C |
F |
GAGAACCAAAGACAAAGACCTGGA |
465 |
60°C |
|
R |
ACGACAAACCTGAATTGCCTCCTT |
|||||
11. |
P |
F |
CTATGGCCCTGCAAACGCGCCAGAAA |
534 |
60°C |
|
R |
TCACGCGCCAGGGCGCAGCC |
|||||
12. |
T |
F |
TTGGCCTGTTTGTGCCTAAACCAT |
750 |
60°C |
|
R |
CGTTGATTACACTTAGCTTTGGAC |
|||||
13. |
K/B |
F |
GCGGTCCGGAAAGCCAGAAAAC |
160 |
60°C |
|
R |
TCTTTCACGAGCCCGCCAAA |
|||||
14. |
W |
F |
CCTAAGAACAACAAAGCCCCCG |
242 |
60°C |
|
R |
GGTGCGCGGCATAGAACCGT |
|||||
15. |
FIIA |
F |
CTGTCGTAAGCTGATGGC |
270 |
60°C |
|
R |
CTCTGCCACAAACTTCAGC |
|||||
16. |
FIA |
F |
CCATGCTGGTTCTAGAGAAGGTG |
462 |
60°C |
|
R |
GTATATCCTTACTGGCTTCCGCAG |
|||||
17. |
FIB |
F |
GGAGTTCTGACACACGATTTTCTG |
702 |
60°C |
|
R |
CTCCCGTCGCTTCAGGGCATT |
|||||
18. |
Y |
F |
AATTCAAACAACACTGTGCAGCCTG |
765 |
60°C |
|
R |
GCGAGAATGGACGATTACAAAACTTT |
|||||
19. |
I1 |
F |
CGAAAGCCGGACGGCAGAA |
139 |
60°C |
|
R |
TCGTCGTTCCGCCAAGTTCGT |
|||||
20. |
FreP |
F |
TGATCGTTTAAGGAATTTTG |
270 |
60°C |
|
R |
GAAGATCAGTCACACCATCC |
|||||
21. |
X |
F |
AACCTTAGAGGCTATTTAAGTTGCTGAT |
376 |
60°C |
|
R |
TGAGAGTCAATTTTTATCTCATGTTTTAGC |
|||||
22. |
HI1 |
F |
GGAGCGATGGATTACTTCAGTAC |
471 |
60°C |
|
R |
TGCCGTTTCACCTCGTGAGTA |
|||||
23. |
N |
F |
GTCTAACGAGCTTACCGAAG |
559 |
60°C |
|
R |
GTTTCAACTCTGCCAAGTTC |
|||||
24. |
HI2 |
F |
TTTCTCCTGAGTCACCTGTTAACAC |
644 |
60°C |
|
R |
GGCTCACTACCGTTGTCATCCT |
|||||
25. |
L/M |
F |
GGATGAAAACTATCAGCATCTGAAG |
785 |
60°C |
|
R |
CTGCAGGGGCGATTCTTTAGG |
|||||
26. |
mdh36 |
F |
GTTTAACGTTAACGCCGGT |
274 |
65°C |
(16) |
R |
GGTAACACCAGAGTGACCA |
|||||
27. |
gyrB47 |
F |
CGCGATAAGCGCGAC |
132 |
60°C |
|
R |
ACCGTCTTTTTCGGTGGAA |
|||||






































